This is an AI for Health MSc project. Students are eligible to receive a monthly reimbursement of €500,- for a period of six months. For more information please read the requirements.
Clinical Problem
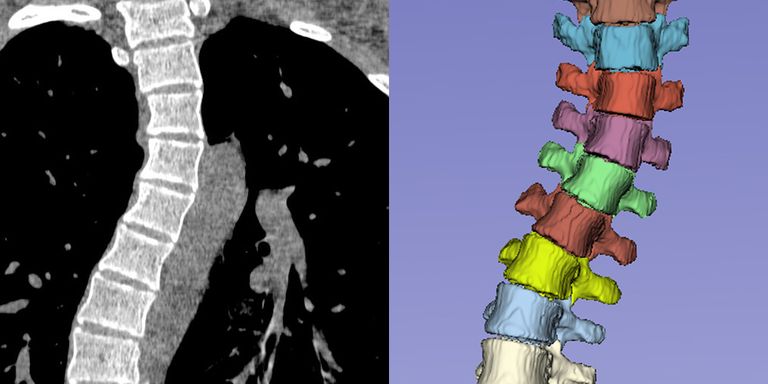
Segmentation and anatomical labeling of the vertebrae is an essential step in computerized analysis of the spine using CT and MR images. The goal of this analysis can be, for instance, detection of osteoporotic compression fractures of the vertebrae, measurement of spinal abnormalities related to chronic low back pain, or planning of surgical correction of idiopathic scoliosis. However, existing training data for automatic vertebra segmentation is limited to relatively normal spine shapes. In clinical practice, spinal deformities present therefore a serious challenge for automatic image analysis algorithms. The goal of this project is to improve the robustness to spinal deformities and thus bring automatic spinal analysis one step closer to clinical practice.
Solution
The project aims to model spinal deformities and develop a simulation framework for generating abnormal but realistic spine shapes. Known normal spine shapes can be used as starting point to develop a parametric graph model of the spine. Using values from the literature, plausible deformations can be defined, e.g., deformation of a normal spine into a S-curved spine with realistic curvature. In order to use these modeled spine shapes to improve the robustness of segmentation algorithms, the synthetic shapes can be used to create a corresponding deformation field and deform CT and MR images of normal spines together with their (already available) segmentations. This synthetic data can then be added to the training data of a learning-based segmentation method to increase the robustness to spinal deformities (data augmentation). The project can focus on the simulation/modeling aspect by using an existing spine segmentation method.
Data
Public and private datasets with more than 400 CT scans and around 100 MR scans are available, together with reference segmentations and anatomical labels of the vertebrae. Additionally, at least 20 scans of patients with scoliotic spines are available for validation of the results. More will be collected from our PACS as part of the project.
Results
The main result will be a model of spinal deformities, which can be used both to generate synthetic images with deformities but which can also be used in other spinal analysis projects to simulate different spine shapes. Practically, the project will also lead to an improved segmentation algorithm, in particular for scans of patients with scoliosis. Here, this algorithm can help plan surgical interventions and design patient-specific drilling templates. The final deliverable is a code repository on GitHub and a publicly available algorithm on the platform Grand Challenge.
Embedding
The student will be supervised by a research scientist from the Diagnostic Image Analysis Group, with expertise in deep learning and in particular spine segmentation and analysis in medical images, and by a research scientist from the Department of Orthopedics, with expertise in musculoskeletal modeling. An orthopedic surgeon will provide clinical expertise. For model development, we provide access to a large GPU computation cluster and an existing database of spine scans and reference annotations.
Requirements
- Students Artificial Intelligence, Data Science, Computer Science, Bioinformatics, Biomedical Engineering or similar in the final stages of their Master education.
- You should be proficient in Python programming and have a theoretical understanding of deep learning architectures.
- Experience with medical images and with programming in MATLAB is beneficial.
Information
- Project duration: 6 months
- Location: Radboud University Medical Center
- For more information or to apply for this project, please contact Nikolas Lessmann.



