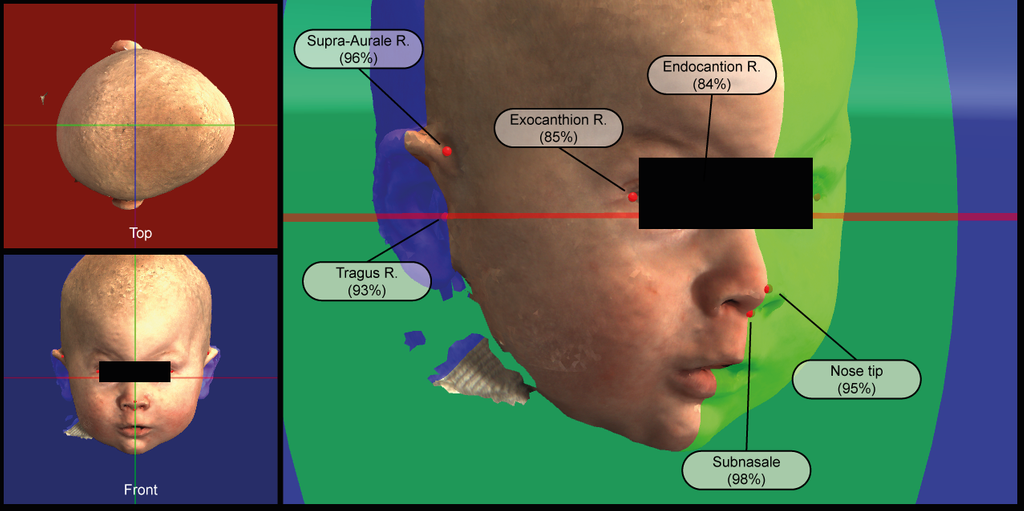
Clinical Problem
Three dimensional (3D) landmarks are used in various fields within medicine. The 3D landmarks are often used in the alignment of 3D radiological images, 3D scans or 3D photos. Other common applications are object recognition and tracking. The placement of 3D landmarks is used in the Radboudumc 3D-Lab and the department of Oral and Maxillofacial Surgery for surgical planning, follow-up and diagnostics. Manual placement of 3D landmarks is a cumbersome process with a high degree of inter- and interobserver variability depending on the task at hand. Accurate and consistent landmark placement is key in the planning and follow-up of 3D imaging with landmark based alignment. The results of automated 3D landmark placement largely vary on the capture device/data source, pre-processing, sampling resolution, algorithm, etc. Automation of 3D landmark placement with the use of artificial intelligence is less straightforward compared to landmark placement in volumetric or 2D radiological images. In contrast to the landmarks in volumetric radiological images, the landmarks in 3D imaging are not based upon coordinates within a volumetric matrix but rather a set of semi- or unordered coordinates in 3D space. However, there is great need for accurate, objective and automated 3D landmark detection with the Radboudumc 3D-Lab and the department of Oral and Maxillofacial Surgery for the numerous of tasks with 3D data in these departments.
Solution
We would like to have 3D landmark detection based upon the 4 most common 3D imaging modalities within our departments while fully or partially using artificial intelligence (AI). With the recent successes and the emerging field of AI within 3D technology we strongly believe that AI can outperform manual and other (semi-)automated 3D landmark detection solutions both in time and accuracy. Within this project we aim for 3D landmark detection of the face and/or cranium in one of the following modalities: bony-tissue CT-scans, soft-tissue CT-scans, soft-tissue 3D photos and soft-tissue textured 3D photos. Per modality a set of 12 to 36 clinically relevant landmarks have to be detected.
Data
We have access to several hundreds of annotated CT-scans and 3D Photos with varying number of landmarks from retrospective studies from our departments. Depending on the modality the most suitable database will be used.
Approach
Students will be supervised by a 3D Lab research member whose research is dedicated to 3D technology and machine learning techniques. One modality will be chosen at the start of the project. Depending on the chosen modality a combination of convolutional neural networks (CNNs) and/or feed-forward neural networks can be used with or without a combination with more conventional 3D processing algorithms. The goal is accurate, objective and automated 3D landmark detection of the face and/or cranium in the chosen modality. The final deliverable will be implemented within the 3D-Lab software suite called 3DMedX. Furthermore, this final deliverable will also be a publicly available algorithm on the platform Grand Challenge.
Requirements
- Students with a major in data science, computer science, or artificial intelligence in the final stage of master level studies are invited to apply.
- Interest in (medical) image analysis and machine learning.
- Affinity with programming in Python and with deep learning packages (e.g. PyTorch or Keras) is required.
Information
- Project duration: 6 months
- Location: Radboud University Medical Center
- For more information, please contact Guido de Jong



